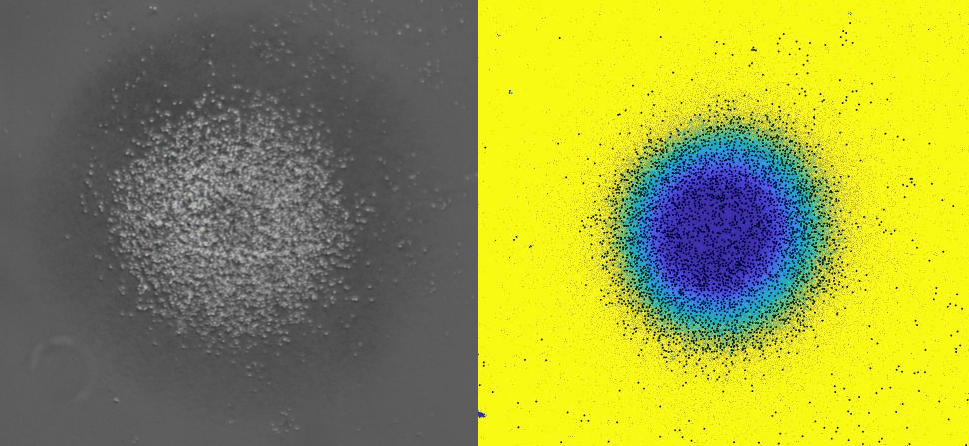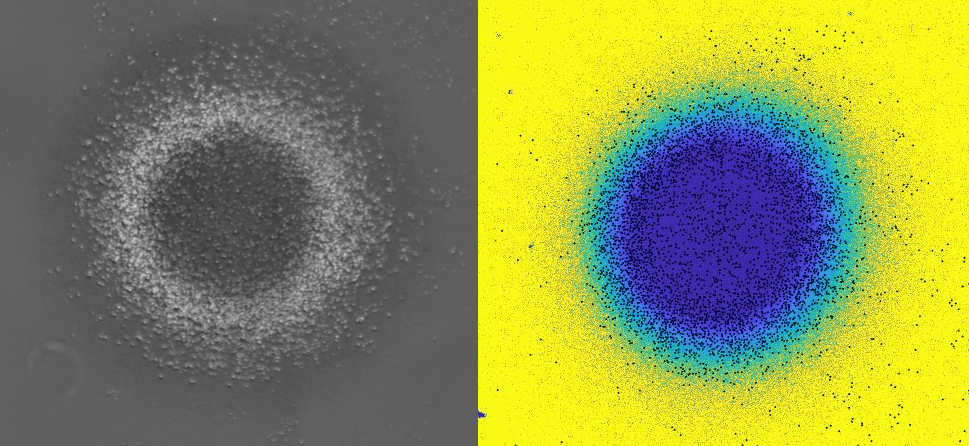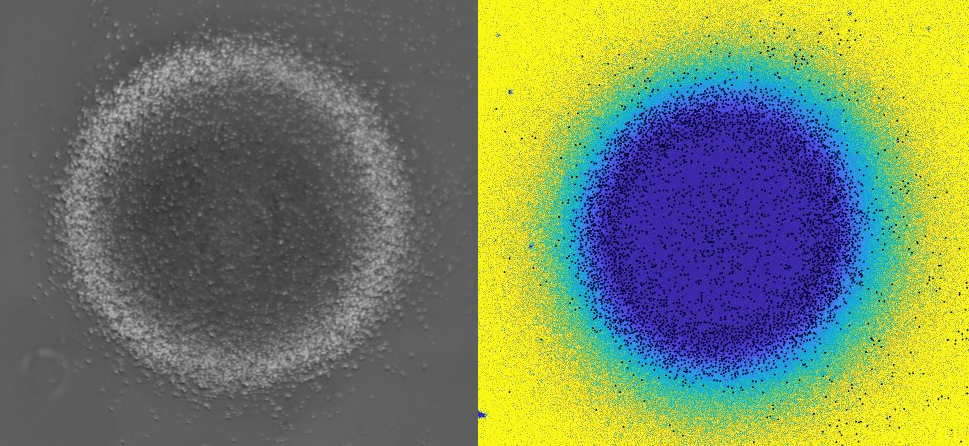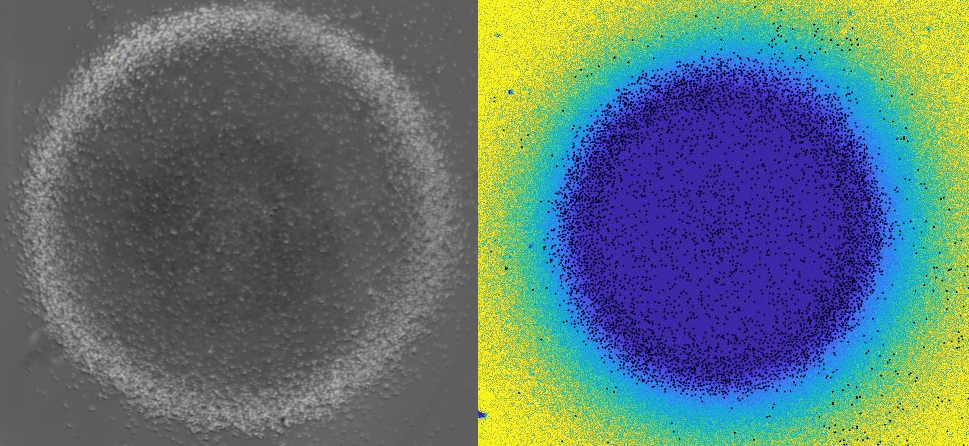From cell-cell interactions to emergent collective dynamicsChristophe Anjard, Olivier Cochet-Escartin, Hélène Delanoë-Ayari, Jean-Paul Rieu, Charlotte Rivière Using various model systems and common videomicroscopy and image analysis tools, we study the dynamics of large assemblies of cells in interaction and we do so in a variety of contexts: epithelial-mesenchymal transition, cardiac assay, population dispersion and swarming… While in vitro, our assays allow to study the complexity of multicellular assemblies in controlled environments (confinement, patterning, gradients of oxygen and other chemical factors). At low to intermediate densities, cells migrate individually but can bias their random trajectories by various short and long range interactions. Near confluence, cell monolayers exhibit transitions between states of convective motion with various correlation lengths. These observations require the development of multiscale models. Fundings : ANR, MITI (CNRS). Collaborations: MSC Paris, Porto Alegre (Brasil), Diag'n Cell Montpellier, West's Lab (University Georgia, USA), IFS Tohoku (Japan), ICJ Lyon, IBENS Paris. The motility of the social amoeba dicty
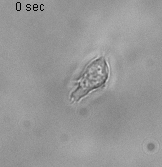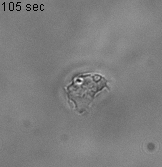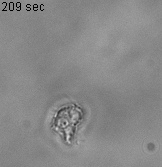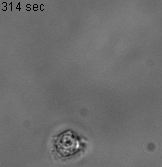
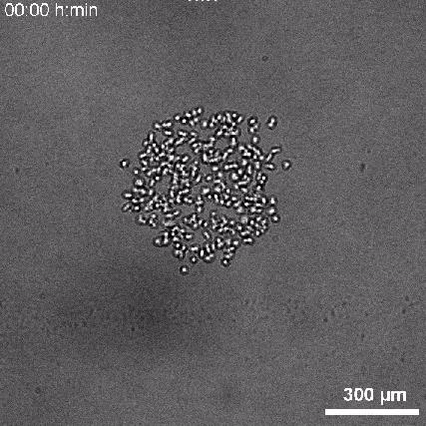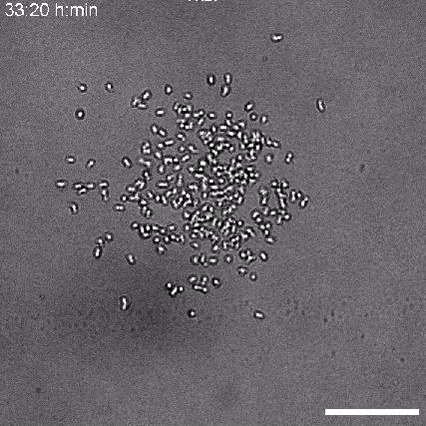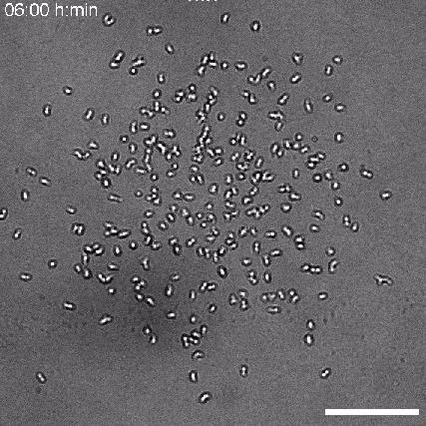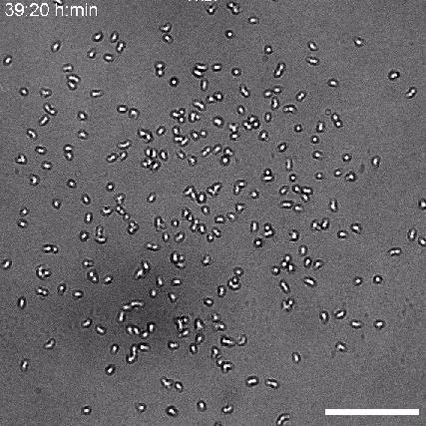
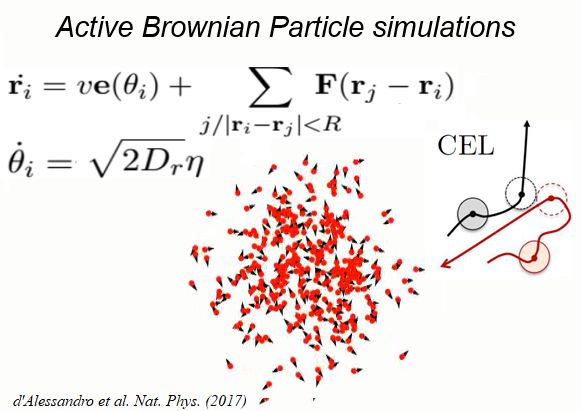
Using 2D motility assays, we have shown that the migration of vegetative dicty cells is regulated by secreted quorum sensing factors (QSF) [1]. From newly developed 2D spreading assay using PDMS stencils, we discovered that the spreading of microcolonies is faster when cell density increases (Fig. 1). At long times, the accumulation of secreted QSF will act to reduce the overall cell motions, thus preventing them to drift too far apart. But at short times, cell-cell collisions induce Contact Enhancement of Locomotion (CEL). These observations look closely related to the concept of ‘Contact Inhibition of Locomotion’ (CIL) described long time ago [2] and recently thoroughly investigated in various cell lines [3]. A deeper understanding of the spreading of dicty, including using simulations and theoretical modelling could pave the way towards a better global understanding of contact interactions in between individual cells.
Collective behavior of Dictyostelium discoideum.We are now investigating the response of an assembly of Dicty cells to oxygen gradients and hypoxia. To do so, we develop microsystems allowing to tightly control oxygen distributions. We also manufacture oxygen sensors which can be embedded in our experiments to observe, in one setting, both the distribution of oxygen and the behavior of the cells. These experiments are complemented with the screening of mutants showing defects in aerotaxis (collaboration Prof. West, University of Georgia), our own numerical simulations using cellular Potts models and mathemical modelling of collective responses by kinetic models (collaboration Prof. Calvez, UCBL). This work provides us with a framework, both experimental and theoretical, to study hypoxia and self-generated gradients on many different systems. Therefore, we plan to extend our results to different biological situations. For example, oxygen is thought to be important in guiding neutrophils during an immune response but a systematic study of the aerotaxis of such cells is still lacking.
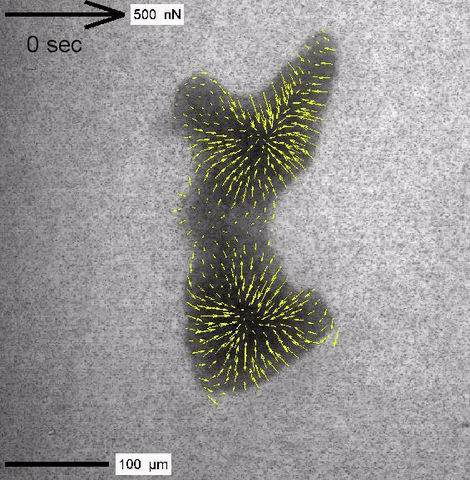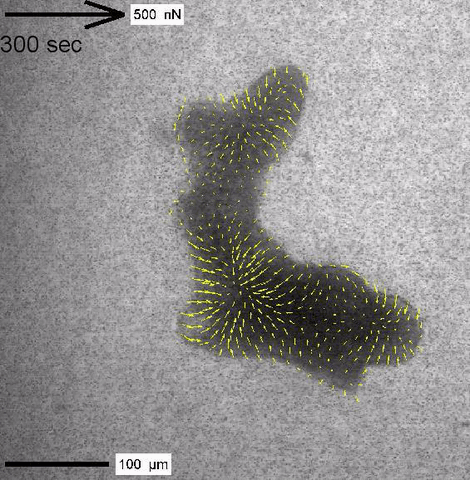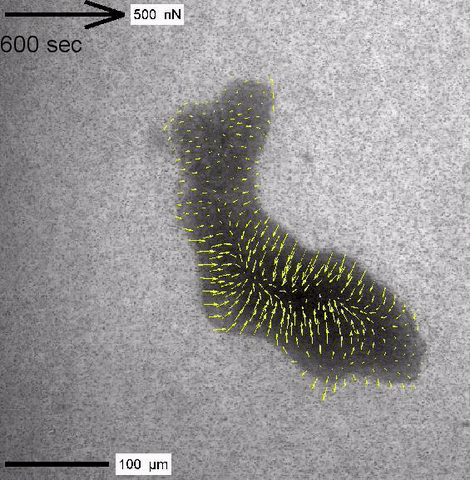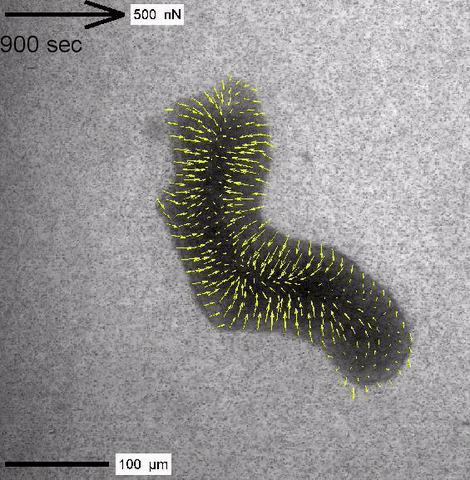
Cytoplasmic streaming and friction forces
|