Technological developments
Image analysis by deep or machine learning
We develop and use deep learning approaches in order to analyze images stemming from our experiments. In recent years, machine learning has proved to be a very powerful and versatile tool in image analysis but these approaches require technical knowledge which is often remote from biological expertise. We thus take advantage of our positioning at the interface between physics and biology to develop these models in-house.
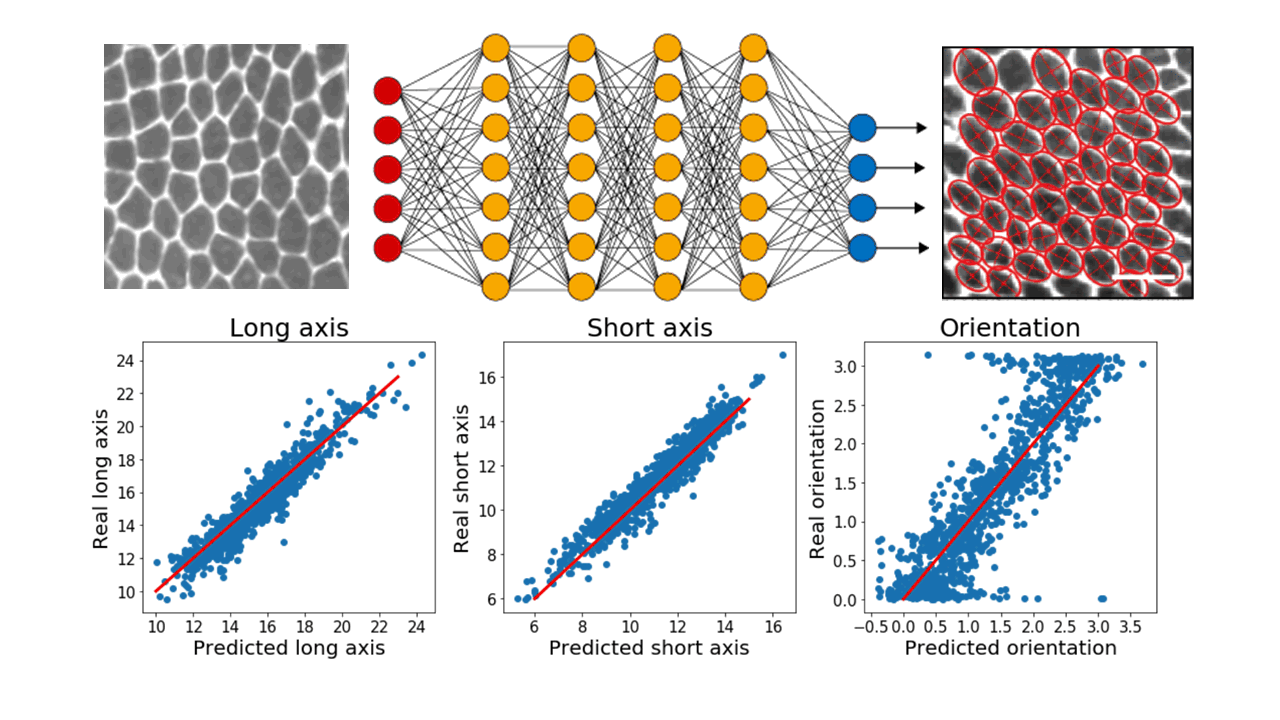
In particular, we make extensive use of convolutional neural networks to extract information from images of cellular monolayers (in 2d) or spheroids (in 3d). Our current models are able, for example, to automatically detect cells and compute relevant morphological measurements such as cell sizes and elongations. Now, we are developing new models to be used for segmenting 3d image stacks and to detect dynamic events in timelapses.
 |
|
Top: raw images of cellular monolayers are passed through a deep neural networks which outputs morphological measurements of the cells. Bottom: Current models properly predict these quantities on a training test annotated by hand. Red lines are a guide for the eye representing perfect agreement between values predicted by the model and real values.
|
Potts
Potts models are discrete spatio-temporal models designed to simulate the behavior of cells in various conditions. They rely on energy terms representing cell adhesion, motility, chemotaxis etc… and Monte Carlo simulations to converge towards minimal energy configurations.
The versatility of this tools allows to simulate complex situations such as, for example, the collective response of Dictyostelium cells to hypoxia. This phenomenon depends on the diffusion and consumption of oxygen by cells, their different behavior depending on local oxygen concentration (aerokinesis), their response to oxygen gradients (aerotaxis), their division rate and their unbiased motility. All these ingredients can be implemented in a Potts model using CompuCell3D and Python as programming languages.
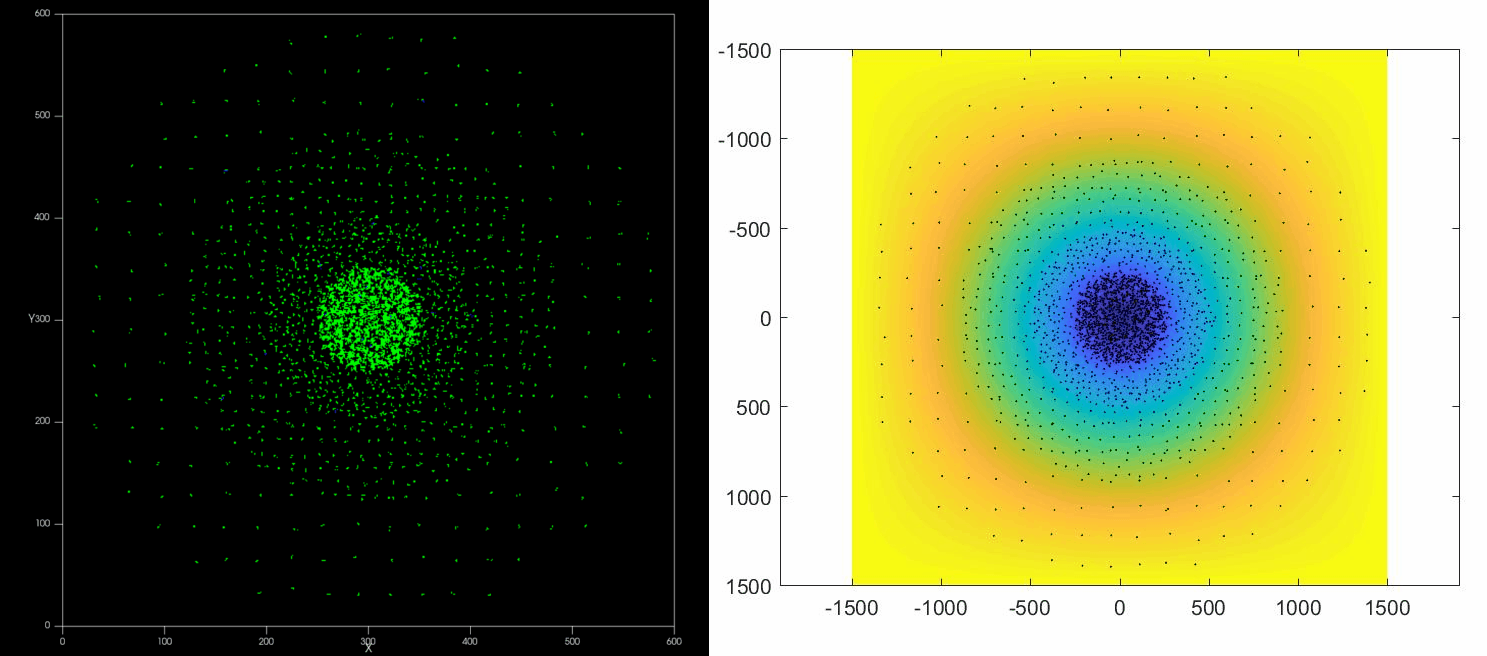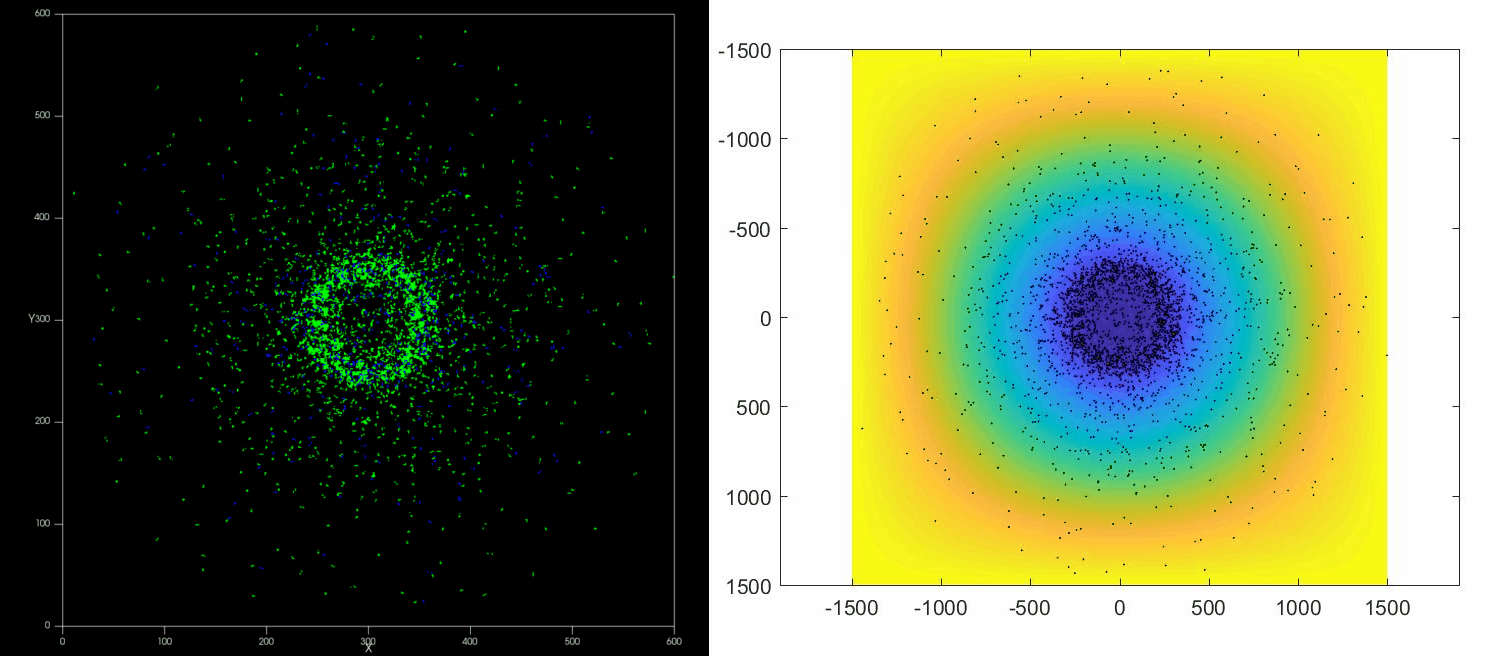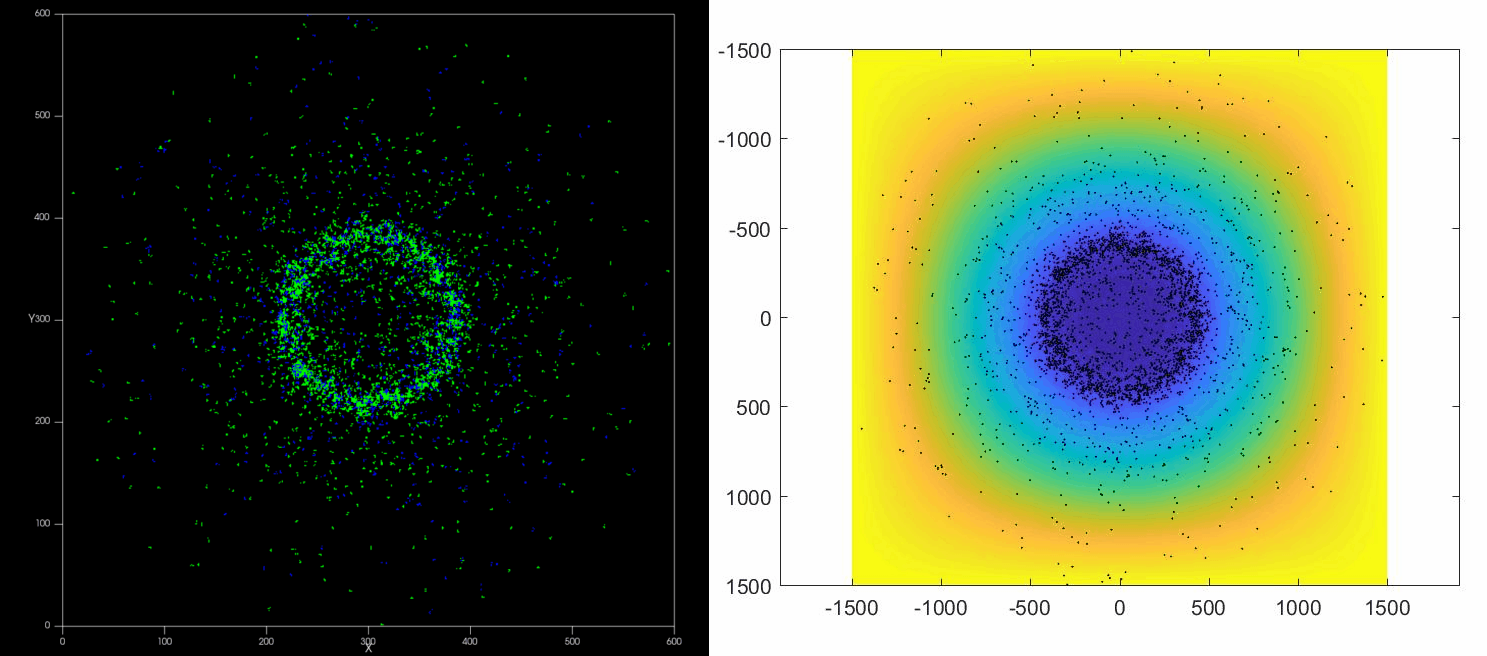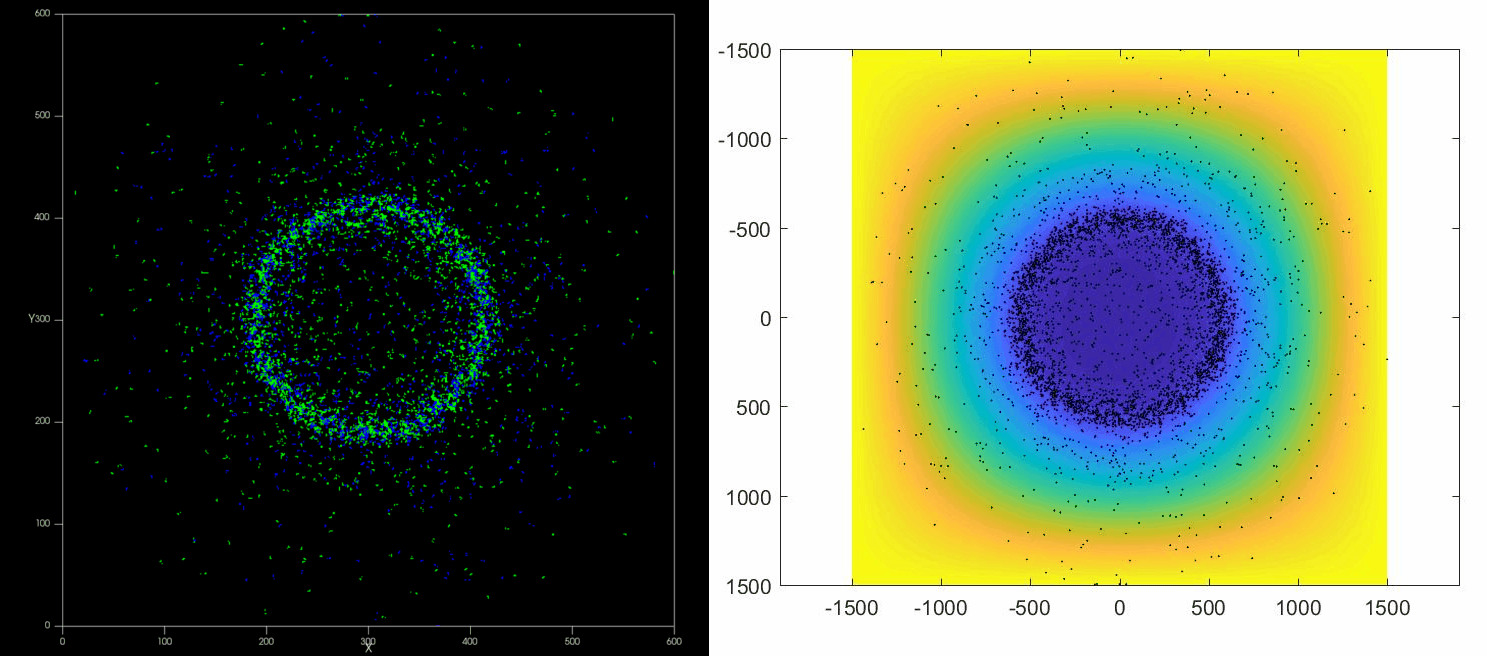 |
| Left: Potts simulation of the behavior of a spot of Dictyostelium cells under self-generated hypoxia. Right: reconstruction of the cell motion (black circles) and oxygen profiles (background colormap) stemming from the numerical simulations |
Optoacoustic and Brillouin imaging
 |
Optoacoustic techniques allow generating and detecting sound waves remotely with lasers. With frequencies ranging from a few MHz to tens of GHz, they allow monitoring the ultrashort timescale mechanical response of the sample. We use these techniques to probe the mechanical properties (stiffness and viscosity) of cells and tissues with a micrometer resolution. |
|
We also have developped a life science microscope based on a Brillouin spectrometer that allows investigating the dynamics of tissues over several days. Brillouin microscopy is label-free and offers a contrast based on the mechanical and optical properties. We have used it to investigate the role of mechanics in the resistance of tumors to therapy, and to analyze mineralized tissues.
For more information: BLS movie.
|
 |
Microfabrication / Microfluidics:
We develop microfabricated and microfluidic tools to apply constraints, control cell and tissue environment or measure biophysical parameters. Such devices can be produced with soft lithography techniques (INL Clean room facility), micromilling or 3D printing available in the lab. Devices can be classically fabricated in PDMS (Polydimethylsiloxan), but we have also developed hydrogel-based (agarose) approaches.
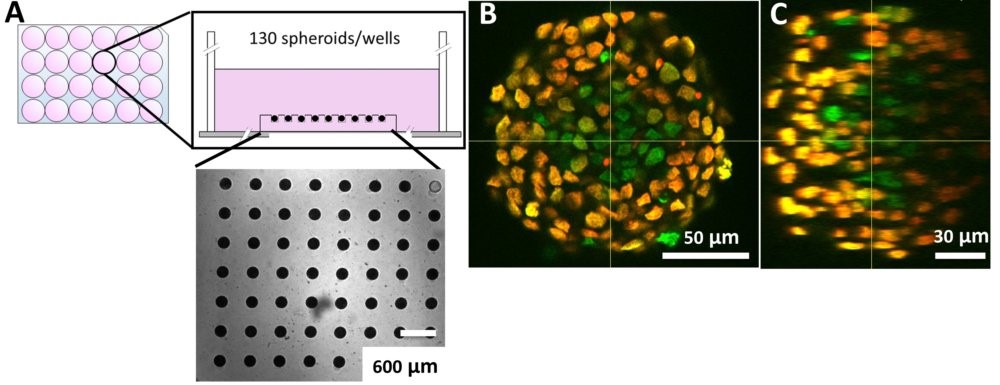
For example, we have developed a flexible agarose-based microsystem that precisely confines cells for extended periods. This innovative approach faithfully reproduces long-term in vitro confinement mimicking in vivo conditions, without affecting cell behavior by other means. This is a unique tool to analyze the role of long-term effect of mechanical confinement in normal and pathological conditions.
|
Measure cellular volume throught time: FXm
We also use microfabricated chamber to measure single cell volume adaptation to osmotic or mechanical stresses. This technique of exclusion of fluorescence originally developed in the team of M. Piel allows assessing the dynamics of tens of single cells with excellent accuracy.
|
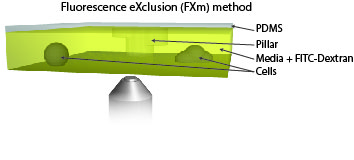 |
We also developed microfluidic devices to apply a large variety of constraints (confinement, shear or osmotic stresses).
4D Traction force microscopy
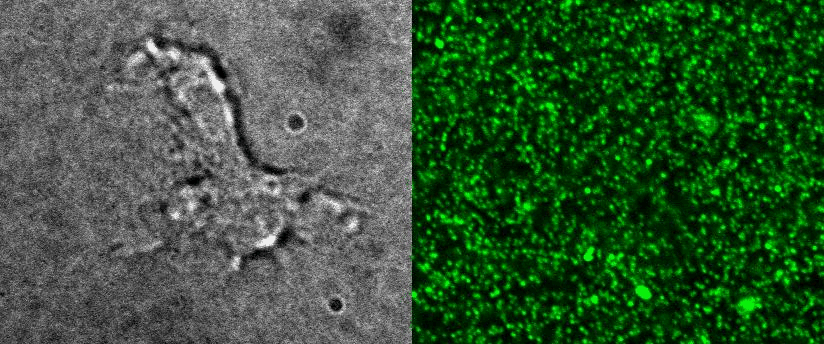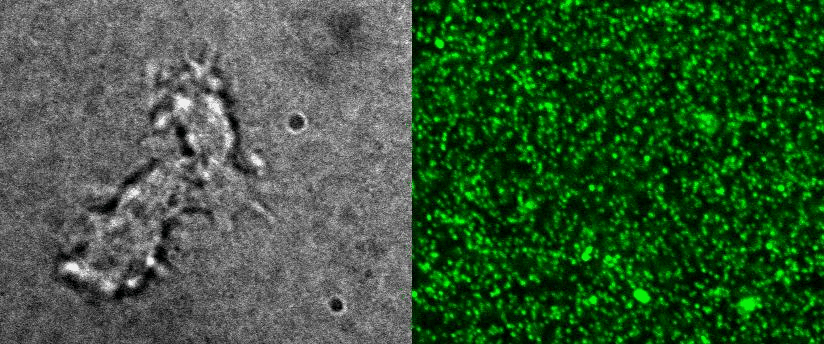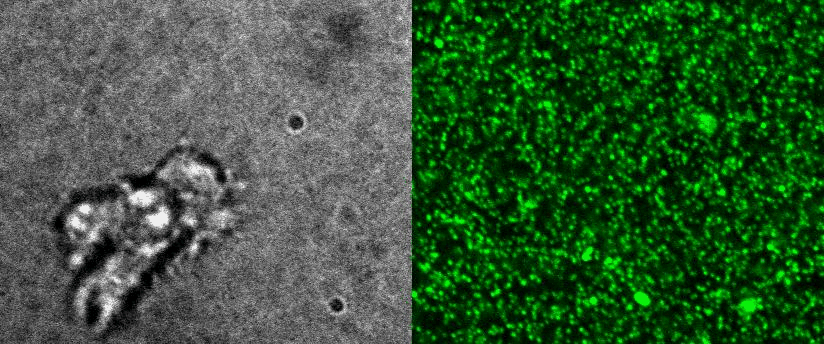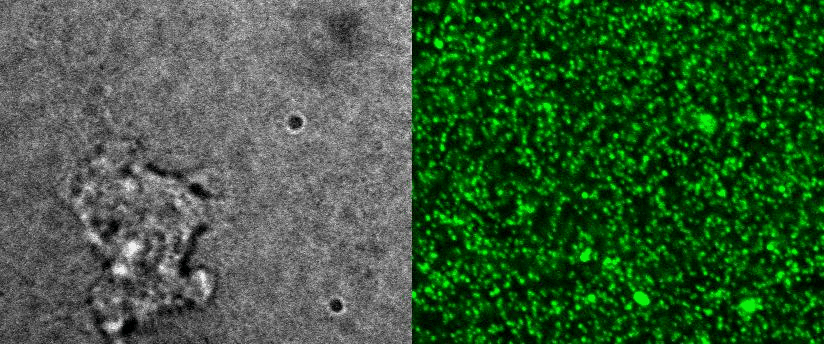 |
To better understand cell motility in 2D, we have developed measurements of the forces exerted by cells on elastic substrates. For this, we insert fluorescent nanomarkers inside polyacrylamide substrates (whose Young Modulus falls between a few hundred Pascals to a few kiloPascals). When cells moved on the substrate we will monitor the displacement of the markers using 3D timelapse microscopy (See movie). Once the displacement is obtained, the forces exerted by the cells can be calculated back using laws of linear elasticity in semi-infinite mediums. |
In-situ immunostaining and imaging of spheroids
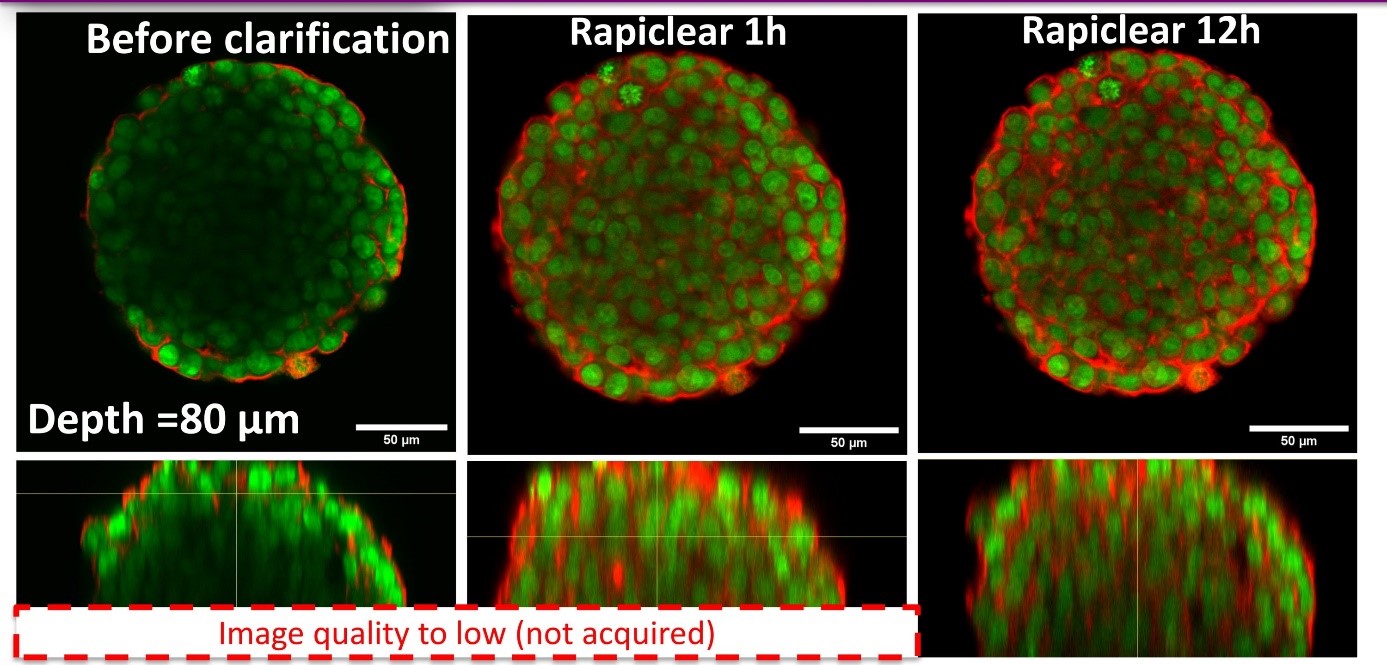
Using our hydrogel-based microsystems, we have developed a user-friendly technique for in-situ immunostaining and imaging of 3D samples such as spheroids or organoids. Our approach is fully compatible with passive and efficient clarification technique, allowing the 3D image acquisition of all-mount spheroids using a classical confocal set-up (up to a depth of 100 µm, see Figures).
Molecular biology:
Most of our work questions the interplay between physics and biology and our ambition is to include both dimensions in our studies. As such, we need to use biological techniques to create new transgenic lines, measure gene expression, visualize expression patterns or even knock down genes via RNA interference. For that purpose, we have equipped a microbiology and molecular biology room for DNA, RNA and protein purification, manipulation, amplification and analysis. We can perform E. coli transformation and culture, PCR, RT-PCR, nucleic acid electrophoresis, gene cloning, protein electrophoresis and western-blot.
|